Convolutional Neural Network for Detecting Cancer Tumors in Microscopic Images
How we won first place at the IdeaLabAi Hackathon hosted by the Sahara Ventures and the Tanzania AI Lab
Cancer of all types is increasing exponentially in the countries and regions at large. Cervical cancer, which is caused by a certain strain of the Human Papillomavirus (HPV), presents a significant public health threat to women on the African continent. All but one of the top 20 countries worldwide with the highest burden of Cervical cancer in 2018 were in Africa in which Tanzania was among.
In Tanzania, cancer control activities and services are undertaken by a wide range of government and non-government agencies, most of which have been done by ORCI and to a lesser extent by some other NGOs. If you need detailed information on this, read more about it here.
AI Commons is a nonprofit organization supported by the ecosystem of AI practitioners, entrepreneurs, academia, NGOs, AI industry players, and organizations/individuals focused on the common good. The organization has gathered the best minds in AI to support the creation of a knowledge hub in AI that can be accessible by anyone, that can help inform governance, policymaking, and investments around the deployment of AI solutions, and be a catalyst for supporting diversity and inclusivity in how AI is deployed for sustainable development goals.
The AI Commons Health & Wellbeing Hackathon is an online competition to solve identified local health problems in Tanzania utilizing AI in which each team consisted of 5 participants.
How It Started
As a participant who was in search of team members connected with Alfaxad Eyembe, a Backend Developer and AI Enthusiast, after which notions and ideas were brought down after we brainstormed.
We concluded on building a model comprising of Deep Convolutional Neural Networks(CNN) and a Web App that screens microscopic images so as to detect cancer tumors, thus increasing the speed, accuracy in cancer diagnosis and testing.
Anthony Mipawa, a Software Engineer, Sang’udi E Sang’udi, a UI/UX Designer, Salome Rumold Mosha, the Pitcher, all of which are Tanzanians, were the other members of the team.
I was the only Nigerian, also the only person to take up the Machine Learning Engineer role since our solution wrapped around building a CNN model for image classification.
Solution Implementation
- Introduction
- Dataset
- Data Preprocessing
- Methodology
- Evaluation
- Deployment
Introduction
Full digitalization of the microscopic evaluation of stained tissue sections in histopathology has become feasible recently due to the advances in slide scanning technology and reduction in digital storage cost. It also increased the computer-aided diagnostic which was why we chose microscopic images to x-ray images as x-ray images will always appear as shades of grey.
Left to do was to get the tons of microscopic images needed for training and making predictions by classifying future images if they contain cancer tumors or not. However, while researching on different health data platforms, I came across the PatchCamelyon(PCam) benchmark dataset.
Dataset
The PCam is a new and challenging image classification dataset that consists of 327.680 color images (96px, 96px) extracted from histopathologic scans of lymph node sections with each image annotated with a binary label indicating the presence of metastatic tissue.
However, the original PCam dataset contains duplicate images due to its probabilistic sampling. Then luckily, I saw a competition hosted on Kaggle to challenge researchers to create an algorithm that identifies metastatic cancer in small image patches taken from larger digital pathology scans, which is a slightly modified version of the PCam dataset.
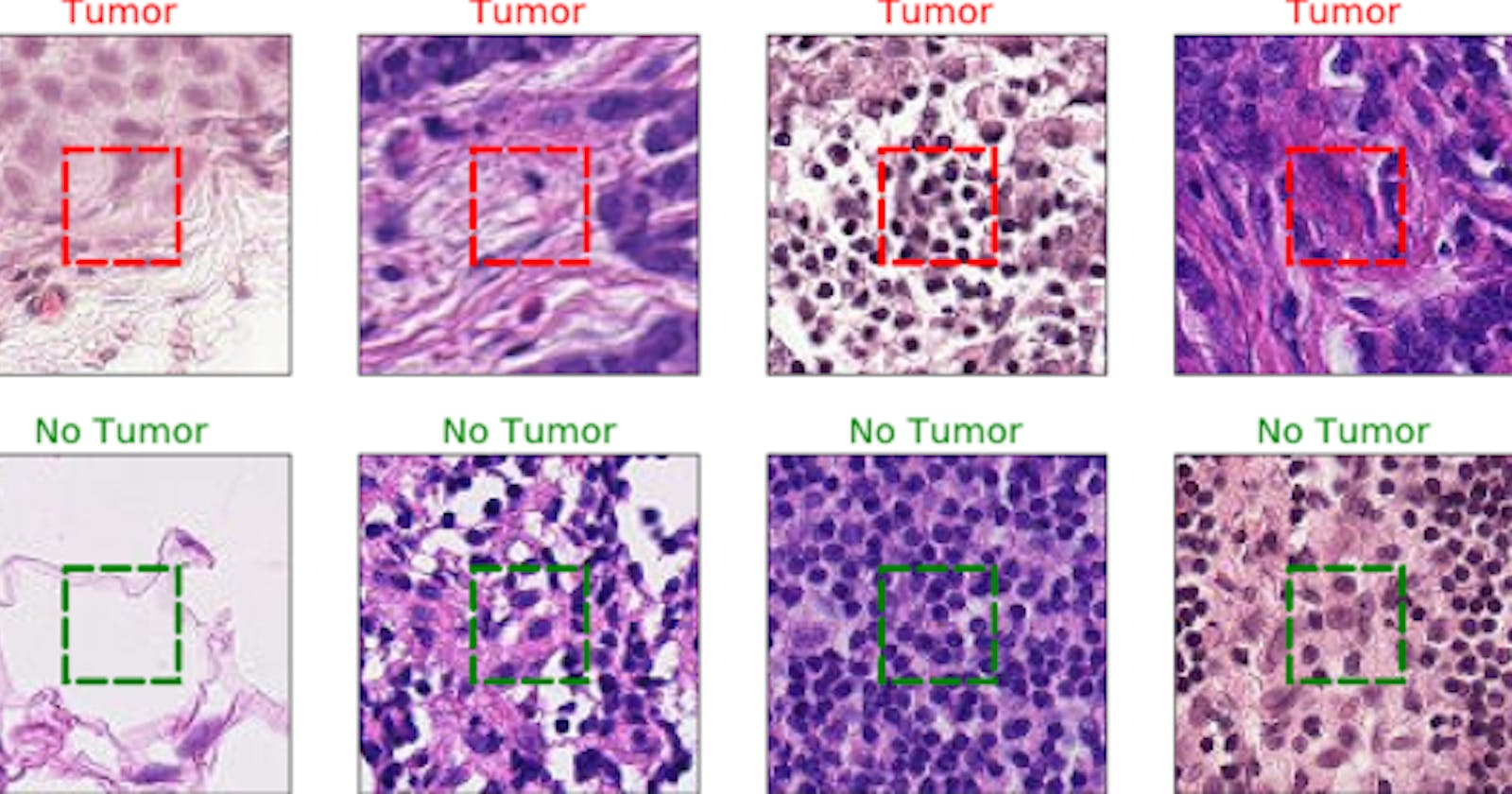
The Kaggle modified one consists of 220,000 training images and 57,458 test images while all images are 96×96 pixels in size. The challenge was to predict the labels(Tumor: Positive, No Tumor: Negative) for the test. A positive label indicates that the center 32x32px region of a patch contains at least one pixel of tumor tissue.
Tumor tissue in the outer region of the patch does not influence the label. This outer region is provided to enable fully-convolutional models that do not use zero-padding, to ensure consistent behavior when applied to a whole-slide image. With pleasure, that was the exact dataset we needed for our model.
Data Preparation
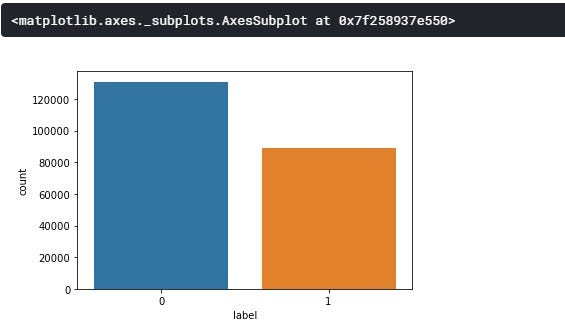
From the class distribution above, we can see that our target values(0 &1) are slightly balanced with class 0(No Tumor) having about 54.16% and class 1(Tumor) with about 45.83% of the whole dataset.
The data preparation raps around using the ImageDataGenerator class from tf.keras.preprocessing.image, a host of Data Augmentation which encompasses a wide range of techniques used to generate “new” training samples from the original ones by applying random jitters and perturbations. Different data augmentation techniques on each image before feeding it to the neural network can be seen below.
- Normalization is a parameter in ImageDataGenerator known as rescale. For most image data, the pixel values are integers with values between 0 and 255. Neural networks process inputs using small weight values, meanwhile, inputs with large integer values can disrupt or slow down the learning process. However, it is good practice to normalize the pixel values so that each pixel value has a value between 0 and 1. So, all we did was to divide all pixel values by 255 which is the largest pixel value.
- Horizontal and Vertical flip, Rotation, Shear, Brightness, Zoom range Applying a tidbit of transformation to an input image will change its appearance slightly, but it does not change the class label.
- Validation Split can separate a portion of your training data into a validation dataset and evaluate the performance of your model on that validation dataset. You can do this by setting the validation_split parameter, most times between 0.2 to 0.5 of your training data. Also, the default value of Batch Size was used(32).
Therefore, data preprocessing/augmentation is a very natural, easy method to apply for computer vision tasks.
Methodology & Evaluation
These are the specific procedures or techniques used to identify, select, process, our proposed solution ranging from architectures, batch normalizations, optimization methods, evaluation methods, and all.
Model Architecture
A Convolutional Neural Network(CNN or ConvNet) is a class of deep neural networks, most commonly applied to analyzing visual imagery. CNN was inspired by biological processes in that the connectivity pattern between neurons resembles the organization of the animal visual cortex. This neural network has one or more layers and it’s mainly for image processing, classification, segmentation, and also for other autocorrelated data. Here goes how the model was compiled:
[Conv2D*3 -> MaxPool2D -> Dropout] x4→ (filters = 16, 32, 64, 96)

Now for the meat of the problem. Four(4) CNN layers were used. We’ll start with a 2D convolution of the image. It’s set up to take 16 windows at first, or “filters”, of each image. CNN is essentially sliding a filter over the input, each filter(kernel size) being 3x3 in size.
The most commonly used Activation Function in neural networks, Rectified Linear Units(ReLU) is used.ReLU for short is a piecewise linear function that will output the input directly if it is positive, otherwise, it will output zero.
Moreso, In CNN, the input layer itself is not a layer, but a tensor. It’s the starting tensor you send to the first hidden layer. This tensor must have the same shape as your training data. For example, we have images of 96x96 pixels in RGB (3 channels), so, this brought about the shape of our input data being (96, 96, 3).
Batch normalization, which increases the stability of a neural network, normalizes the output of a previous activation layer by subtracting the batch mean and dividing by the batch standard deviation is added. We then run a second convolution on top of that with 16, 3x3 windows. A layer that takes the maximum of each 2x2 result to distill the results down into something more manageable is called MaxPooling2D. A Dropout filter is then applied to prevent overfitting.
Not all of these are strictly necessary, you could run without pooling and dropout, but those extra steps help avoid overfitting and help things run faster. These steps are repeated in each of the four layers with the windows being changed within this range 16>32>64>96.
Next, we flattened the 2D layer we have at this stage into a 1D layer. So at this point, we can just pretend we have a traditional multi-layer perception. We then feed that into a hidden, flat layer of 256 units. We then applied dropout again to further prevent overfitting.
And finally, we fed that into our final 1 unit where the softmax function is applied to choose our binary since it exists between 0 and 1. Therefore, it is especially used for models where we have to predict the probability as an output.
Evaluation
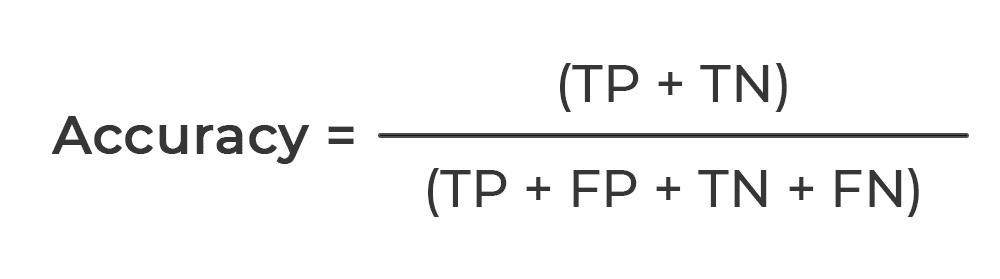
The performance of our proposed classification model was evaluated based on accuracy (ACC). Given the number of True Positives (TP), False Positives (FP), True Negatives (TN), and False Negatives (FN), it’s expressed mathematically as seen below:
Loss Function
Since we have two almost perfectly balanced classes and the accuracy to evaluate the model, we’ve used binary cross-entropy loss function to train our neural networks. The equation for binary cross-entropy loss is:
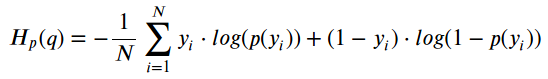
Optimizer
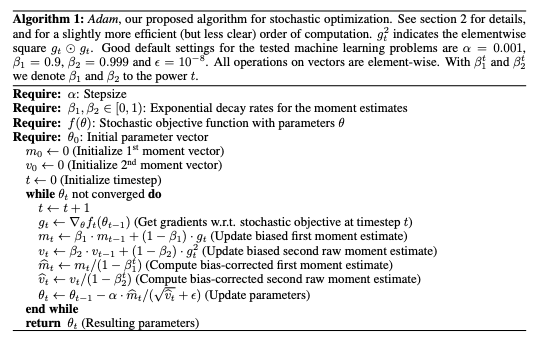
Adam optimizer, similar to vanilla stochastic gradient descent in that it's a first-order, gradient-based algorithm used to optimize stochastic objective functions. It is a cross-breed of the popular RMSProp and AdaGrad optimizers while possessing the attractive properties of both as it works well with sparse gradients and does not require a stationary objective function. It naturally reduces the step size as training proceeds. The algorithm works by keeping track of not only the weight vector but also biased estimates of the first and second moments and the time step. Its pseudo-code can be seen below:
Model Fitting and Training

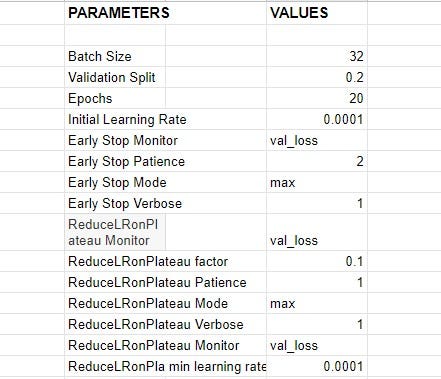
While working towards finding the best parameters for the model, different distinct experiments with various setups were carried out, best setup yielding the top result was selected. The table below shows the list of parameters and their respective values;
Below is a plot showing the training, validation losses, and accuracies of our trained model.
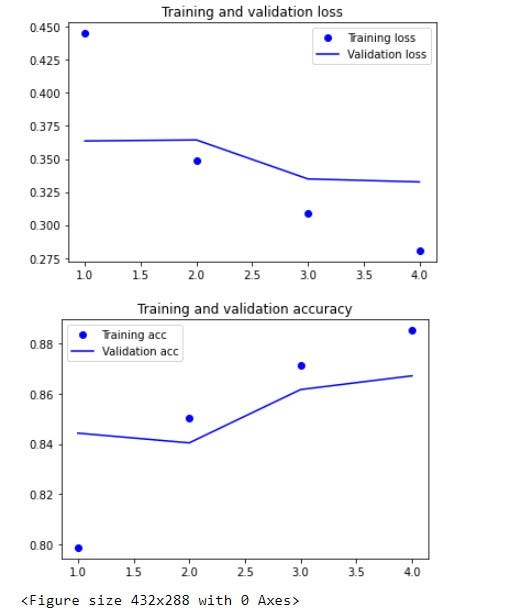
Ultimately, the model was thereafter, saved as a .h5 model for easy deployment into the existing production environment. H5 is a file format to store structured data, it’s not a model by itself. Keras saves models in this format as it can easily store the weights and model configuration in a single file.

Deployment
As Django is written in Python, it makes it a great choice of web frameworks for deploying Machine Learning models. Anthony Mipawa and Alfaxad Eyembe, the software guys among us decided to use Django instead of the Flask framework I’m well versed in. The model was deployed in a web app while Sang’udi E Sang’udi helped in designing the user interface. Pitching and prototype documentation was done by Salome Rumold Mosha who has past experience in such projects and competitions.
Conclusion
We are so proud to provide a novel solution that helps packing the clinically-relevant task of tumor detection into a straight-forward binary image classification task. With an accuracy of 89% on a held-out test set, we are demonstrating the feasibility of this solution. However, our approach can still be improved by using a pre-trained model, ensembling two or three models that tend to outperform single classifiers.
The sole aim of our prototype is to increase the speed, reliability, and accuracy of cancer detection in Tanzania due to the problem of low doctors to patients ratio. Also, exorbitant death of Tanzanian women without even getting diagnosed as a result of Cervical and Breast cancer.
Just today, the winners were announced. And guess what? ELIXIR Won!!
Oh! I forgot to tell you what Elixir is. We named our web app Elixir.
Here are the links you’d like to see;
Click on the clap icon below, share with friends, coding buddies, mentors, and as many people who may need this. More so, you can connect with me on Twitter and LinkedIn in case of questions and more ideas to share. Happy Reading!